The Bootstrap
University of Toronto
July 26, 2023
Motivation
Sampling distributions
Recall: The Central Limit Theorem states that the sampling distribution of the sample mean is approximately normal for large \(n\). That is,
\[ \bar{X}_n \overset{D}{\longrightarrow}Y \;\; \text{where} \;\; Y\sim \text{N}\left( \mu,\frac{\sigma^2}{n} \right) \]
If \(\widehat\mu = \bar{X}_n\) where \(\mu= \mathbb{E}[X_i]\) and \(\sigma^2=Var(X_i)\), then the sampling distribution of is given by the CLT.
\[ \widehat\mu \sim \text{N}\left( \mu, \frac{\sigma^2}{n} \right) \]
Beyond point estimates
So far, we’ve focused on what are called point estimates, our “best guesses” for a parameter value, based on the data we have. But how confident are we in those estimates?
Recall: Estimates are functions of the data, but those functions applied to the random sample are called estimators.
- Unbiased estimators have sampling distributions that are centered on the “true” value.
- The variance of a sampling distribution indicates how accurate an estimate is.
The square root of the variance of an estimator is known as the standard error.
- The standard error is an estimate of the standard deviation of the sampling distribution.
- It is best practices to report the standard error along with any point estimates, in order to provide an indication of accuracy.
Finding sampling distributions
If we know the sampling distribution of an estimator, we can use it for more than finding a estimate.
- What if our estimator is not a sample average?
- Can we make inferences without distributional assumptions?
- Can we estimate the sampling distribution of a general estimator?
The bootstrap method is a way to learn more about the sampling distribution of an estimator, while making fewer assumptions.
The Bootstrap
Illustrations, terminology & notation
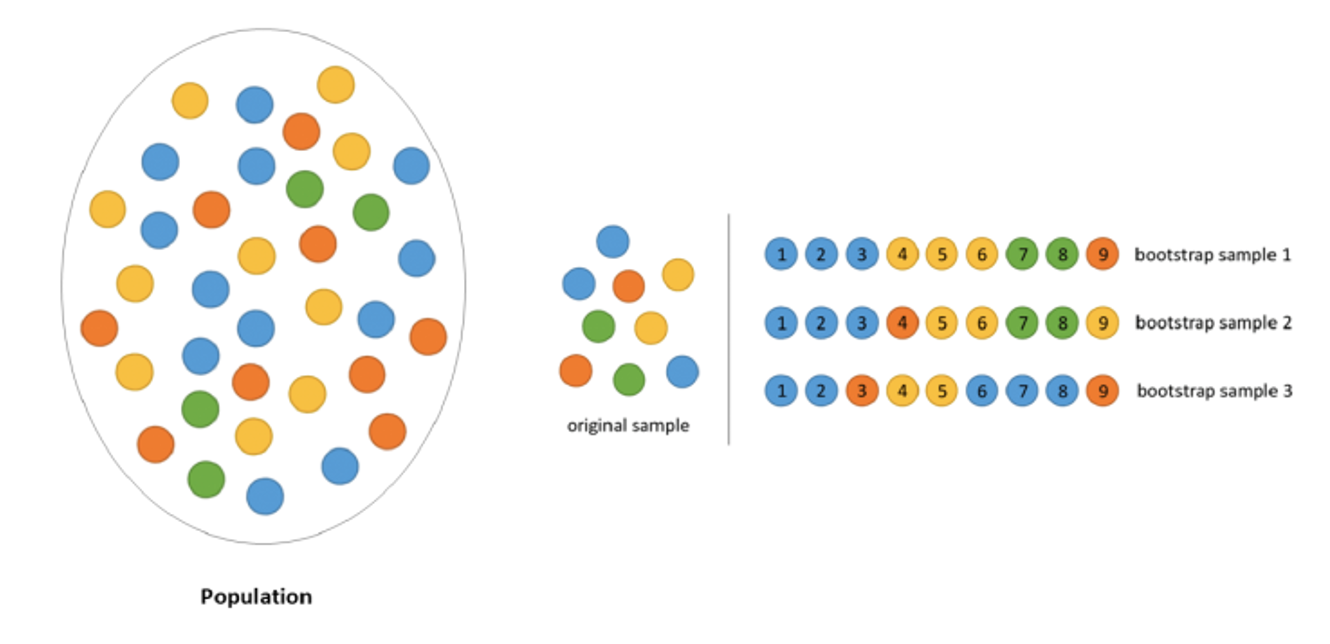
- A bootstrap sample, denoted \(X_1^*,...,X_n^*\), is a sample from a dataset that is drawn with replacement.
- The star notation, \(^*\), is commonly used for to indicate bootstrap samples and estimates from bootstrap samples.
Terminology & notation
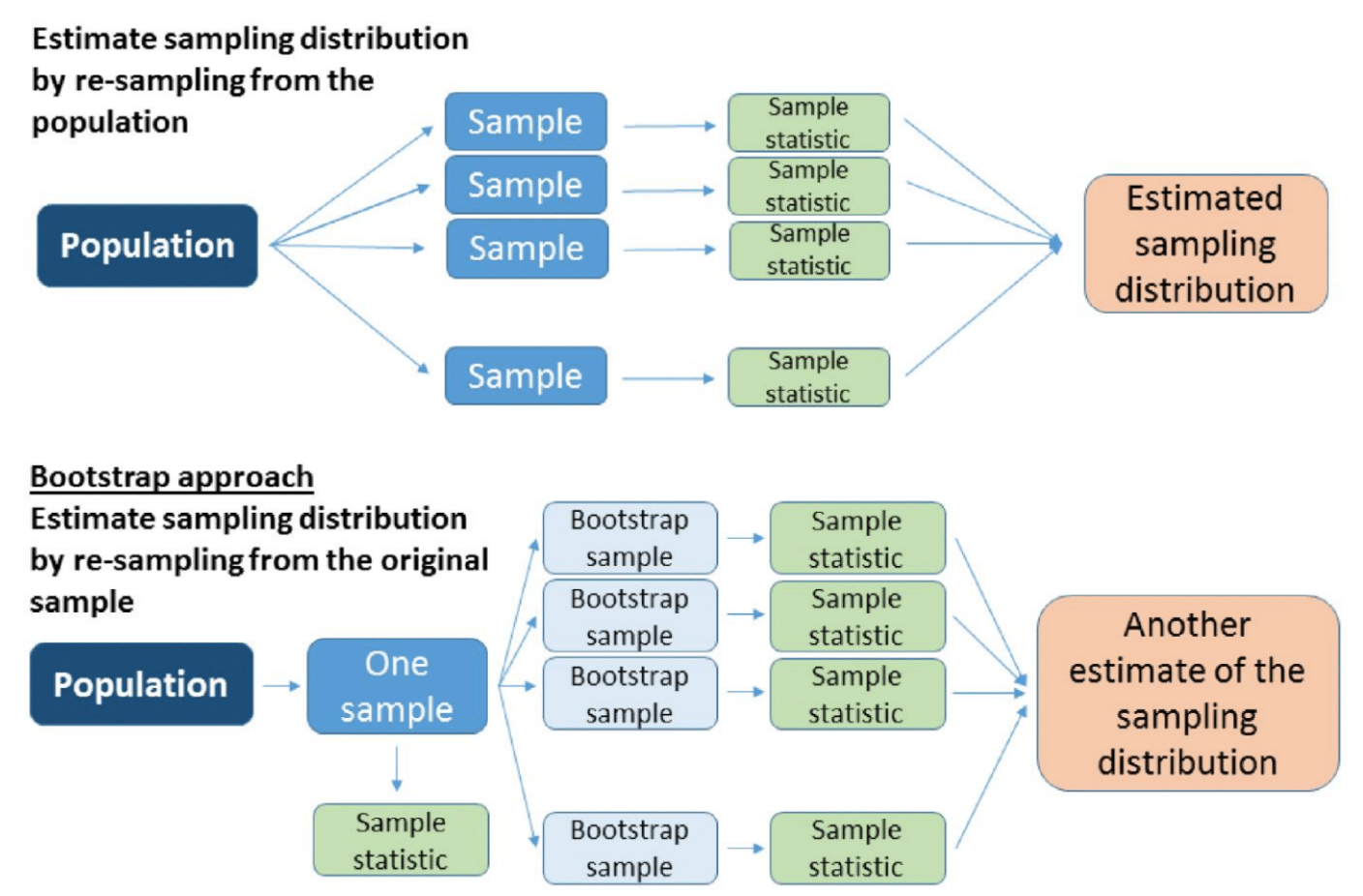
- The estimate \(\widehat\theta_b^*\) is the bootstrap statistic, or bootstrap estimate.
- The distribution of the bootstrap statistic is the bootstrap distribution.
Bootstrap Principle
Approximate the sampling distribution by simulating many datasets from a good model of the distribution of the data and calculate a new estimate of the statistic for each simulated dataset.
- Use the data to compute an estimate of \(\widehat{F}\) of the “true” distribution \(F\).
- Replace the random sample \(X_1,...,X_n\) from \(F\) with a random sample \(X_1^*,...,X_n^*\) from \(\widehat{F}\)
- Approximate the distribution of \(\widehat{\theta}=h(X_1,...,X_n)\) by the distribution of \(\widehat{\theta}_b^*=h(X_1^*,...,X_n^*)\), which is calculated from the random sample.
How to bootstrap
In general
A bootstrap algorithm has the following form:
- By taking a random sample from the original data, generate a bootstrap sample → \(X_1^*,...,X_n^*\)
- Calculate the bootstrap statistic from the bootstrap sample → \(\widehat{\theta}^* =h(X_1^*,...,X_n^*)\)
- Repeat 1. & 2. many times.
Result: Many values of the bootstrap statistic. The distribution of the bootstrap statistic is the bootstrap distribution.
Empirical (or non-parametric) bootstrap
Input
- sample \(X_1,...,X_n\)
- estimator \(\widehat\mu\)
- number of bootstrap samples, \(B\)
Empirical (or non-parametric) bootstrap
Repeat the following for each of \(b=1,...,B\):
- Sample with replacement from \(X_1,...,X_n\) to obtain a bootstrap sample \(X_1^*,...,X_n^*\)
- Compute the bootstrap statistic \(\widehat{\mu}_b^*\) from the bootstrap sample
Empirical (or non-parametric) bootstrap
Output
\(\widehat{\mu}_1^*, ..., \widehat{\mu}_B^*\), which are a sample from the sampling distribution of \(\widehat\mu\)
Why “empirical”?
This algorithm is equivalent to sampling from empirical cumulative distribution function. Recall that the eCDF is defined by
\[ \widehat{F}_n(x)=\frac{\big\lvert \{x_i \vert x_i\leq x \} \big\rvert}{n} \]
Why “non-parametric”?
Notice that no parametric family needs to be specified for the distribution.
Example
“Population”
[1] 6.00168Is that what we expected?
Does it look like a Gamma distribution?
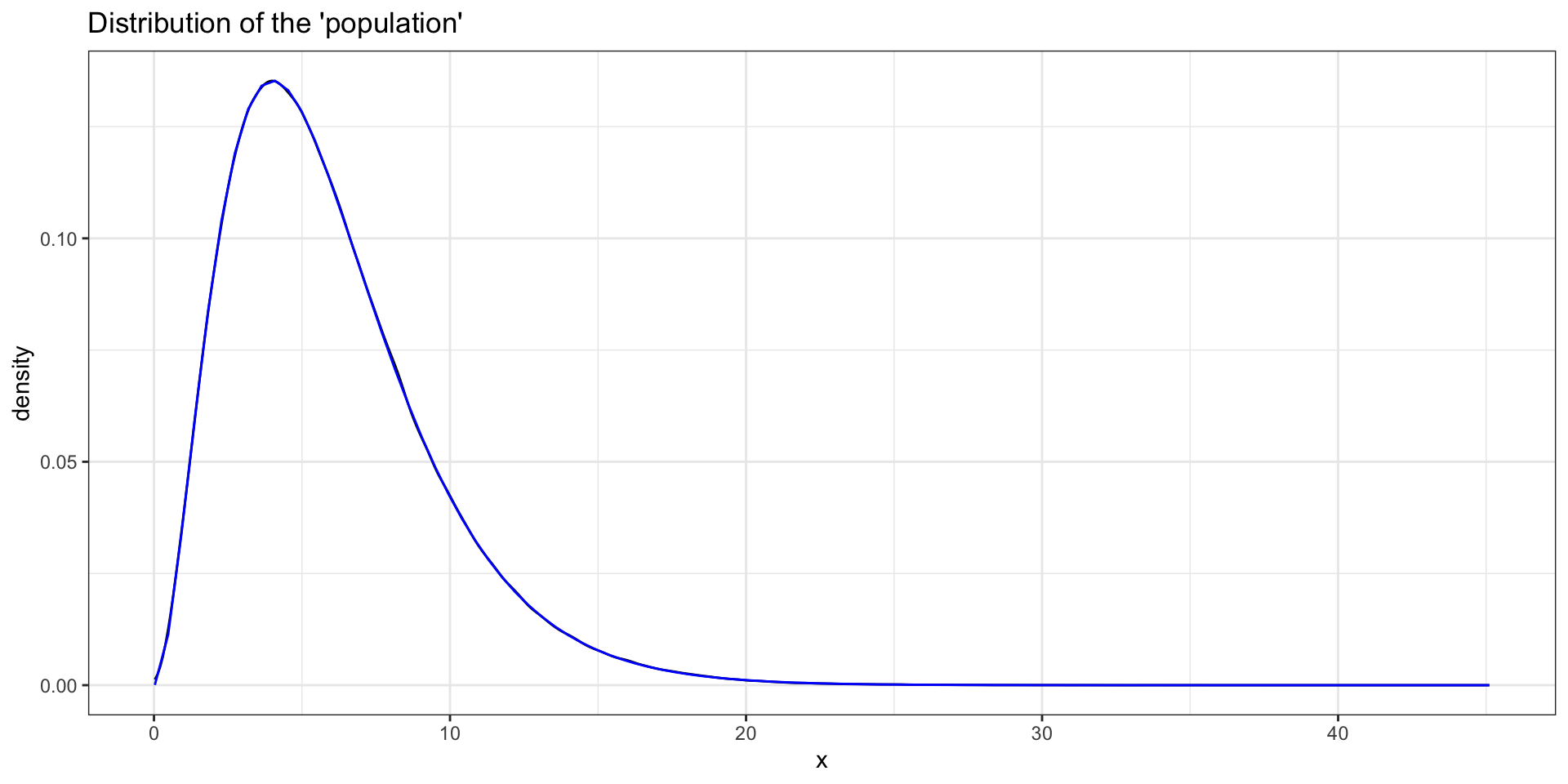
Sample
[1] 6.162984Do we know what the sampling distribution of the sample mean should be for a \(\Gamma\) distribution?
Bootstrap (empirical)
set.seed(238)
B <- 1000 # number of bootstrap samples
n <- length(samp)
bootmeans <- numeric(B) # vector where we'll store B bootstrap means
for (i in 1:B){
bootsamp <- sample(samp, n, replace = TRUE) # sample from the data
bootmeans[i] <- mean(bootsamp) # compute bootstrap stat
}
# output
ggplot(tibble(x = bootmeans), aes(x = x)) +
geom_histogram(aes(y = after_stat(density)), bins = 23, colour = "black", fill = "grey") +
theme_bw() +
labs(title = "Bootstrap distribution of sample means", y = "density") +
geom_vline(aes(xintercept = mu), colour = "blue") +
geom_vline(aes(xintercept = muhat), colour = "purple")Bootstrap (empirical)
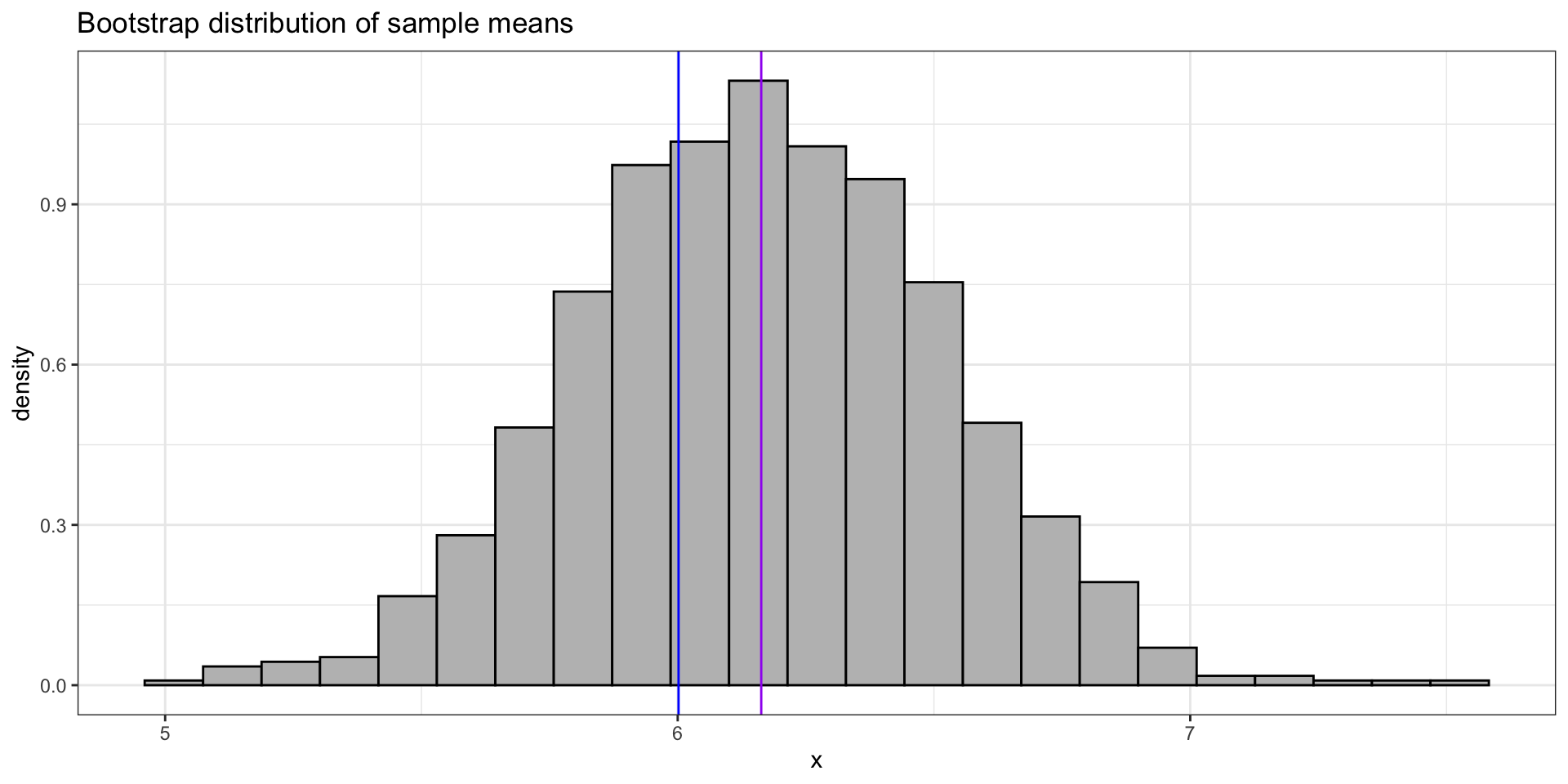
⚠️ The bootstrap does not give a better estimate than the original data because the bootstrap distribution is centered around a statistic calculated from the data. Drawing thousands of bootstrap observations from the original data is not like drawing observations from the underlying population (i.e. the theoretical world). It does not create new data.
How to bootstrap
The bootstrap distribution has approximately the same shape and spread as the sampling distribution, but the center of the bootstrap distribution is the center of the original data (not the center of the theoretical world). So we often use the bootstrap to estimate the sampling distribution of \(\widehat{\theta}-\theta\), the error in our estimate.
Centred estimates
Input
- sample \(X_1,...,X_n\)
- estimate \(\widehat\mu\)
- number of bootstrap samples, \(B\)
Centred estimates
Repeat the following for each of \(b=1,...,B\):
- Sample with replacement from \(X_1,...,X_n\) to obtain a bootstrap sample \(X_1^*,...,X_n^*\)
- Compute the bootstrap statistic \(\widehat{\mu}_b^*\) from the bootstrap sample
- Compute the centered bootstrap statistic \((\widehat{\mu}-\mu)_b^*=\widehat{\mu}_b^*-\widehat{\mu}\)
Centred estimates
Output
\((\widehat{\mu}-\mu)_1^*, ..., (\widehat{\mu}-\mu)_B^*\), which are a sample from the sampling distribution of \((\widehat{\mu}-\mu)\)
We computed \((\widehat{\mu}-\mu)_b^*\) without knowing \(\mu\) (!!)
Example
Bootstrap (centered means)
set.seed(238)
B <- 1000 # number of bootstrap samples
n <- length(samp)
bootcmeans_emp <- 1:B %>%
map(~sample(samp, n, replace = TRUE)) %>% # sample from the data
map(~mean(.x) - muhat) %>% # compute bootstrap stat & center
reduce(c)
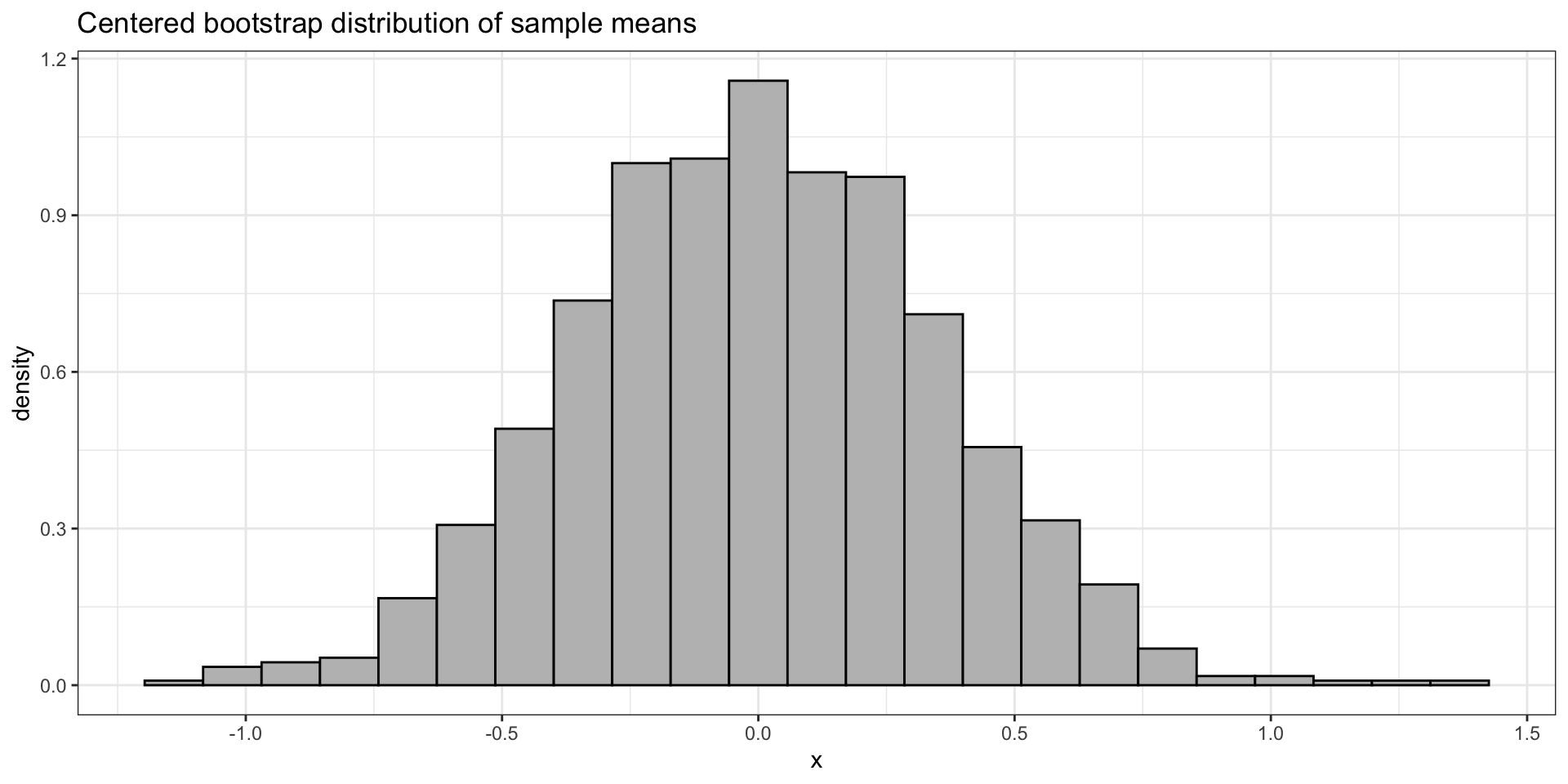
ggplot(tibble(x = bootcmeans_emp), aes(x = x)) +
geom_histogram(aes(y = after_stat(density)), bins = 23, colour = "black", fill = "grey") +
theme_bw() +
labs(title = "Centered bootstrap distribution of sample means", y = "density") In this course, please do not use the boot package or any other similar packages.
Bootstrap (centered means)
How to bootstrap
Suppose we can make a reasonable assumption about the shape of the distribution of \(F\). Even if it’s not totally correct, it allows the bootstrap to be used in situations where it might fail, particularly for smaller sample sizes.
Parametric bootstrap
Input
- sample \(X_1,...,X_n \overset{iid}{\sim}F_\theta\) where \(F\) is known and \(\theta\) is unknown
- estimate \(\widehat\theta\)
- number of bootstrap samples, \(B\)
Parametric bootstrap
Repeat the following for each of \(b=1,...,B\):
- Sample from \(F_\widehat{\theta}\) to obtain a bootstrap sample \(X_1^*,...,X_n^*\overset{iid}{\sim}F_\widehat{\theta}\)
- Compute the bootstrap estimate \(\widehat{\theta}_b^*\) from the bootstrap sample
Parametric bootstrap
Output
\(\widehat{\theta}_1^*, ..., \widehat{\theta}_B^*\), which are a sample from the sampling distribution of \(\widehat\theta\)
Example
Bootstrap (parametric)
set.seed(238)
alphahat <- (mean(samp))^2 / var(samp) # estimate alpha
bootcmeans_par <- numeric(B) # vector where we'll store B bootstrap means
for (i in 1:B){
bootsamp <- rgamma(n, shape = alphahat, rate = beta) # bootstrap sample with alphahat
bootcmeans_par[i] <- mean(bootsamp) - alphahat/beta # bootstrap estimate
}What can we do besides plot the sampling distributions?
Bootstrap (parametric)
Estimate \(\widehat\mu-\mu\):